Digital PCR (dPCR) is an end-point PCR method that offers an alternative to conventional real-time qPCR for absolute quantification of nucleic acids and rare allele detection. The method is similar to qPCR in terms of the reaction components (primers and labelled probe detect sequence specific targets) and the subsequent amplification, but differs in the way the sample target is measured.
In dPCR the nucleic acid sample mix is partitioned into many individual PCR reactions (droplets or wells, see Figure 1). Then amplification of the individual reactions is conducted and each well or droplet will be either positive (1) or negative (0) for the target molecule. The number of positive vs negative reactions is determined and permits the generation of an absolute count of the number of target molecules in the samples.
This simple and reproducible method, does not rely on a calibration curve for sample target quantification, therefore no reference standards or endogenous controls are needed allowing for greater robustness and reproducibility among different labs. Poisson correction is applied to account for individual reactions that may have more than one molecule of target sequence present. The history of dPCR is discussed in the following article: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5129430/ and the technology is reviewed here: https://www.ncbi.nlm.nih.gov/pubmed/29677144.
dPCR is a particularly useful method for detecting rare alleles in cancer and disease, accurately detecting small CNVs and for gene expression analysis of rare mRNAs and miRNAs. It can also be used for accurate NGS library quantification and for pathogen detection and microbiome analysis. dPCR has also recently been widely utilised by several companies for the detection of COVID-19; For example, Bio-Rad has developed a COVID-19 ddPCR assay ‘designed to be a powerful nucleic acid tool in the battle against COVID-19, with sensitivity and precision beyond that of qPCR assays’(https://www.bio-rad.com/featured/en/coronavirus-covid-19-assay-development-vaccine-research.html). ApexBio have also developed a Naica compatible 3-colour Crystal Digital PCR kit which detects two distinct regions of the SARS-CoV-2 positive strand RNA genome as well as simultaneously monitoring PCR effectiveness with an internal reference detecting a human housekeeping gene (https://www.stillatechnologies.com/a-3-color-crystal-digital-pcr-kit-for-detection-of-covid-19/).
In the Genetics Core, we have recently acquired the Naica ™ Crystal Digital PCR™ System from Stilla Technologies which is a High Sensitivity digital PCR Platform.The picture is of me showing off our new machine. You can view the brochure here: (https://www.stillatechnologies.com/wp-content/uploads/2018/10/Brochure_Naica_System.pdf)
In the Naica™ System, digital PCR is integrated into a single consumable, the Sapphire Chips. The sample in each port (4 samples per chip) is partitioned into a 2D array of approx. 30,000 individual droplets of homogenous size by confinement gradient using the Naica Geode. We have 2 of these instruments, so can run up to 24 samples at once. Thermocycling is also performed on chip, in the geode, for each droplet, immediately after crystal generation. Finally the chips are scanned using the Prism3 instrument and Crystal Reader Software using three different fluorescent channels. This allows multiplexing for three different targets per sample. Finally Crystal Miner software automatically measures the concentrations of targeted nucleic acids in each channel and allows visualisation of the data.
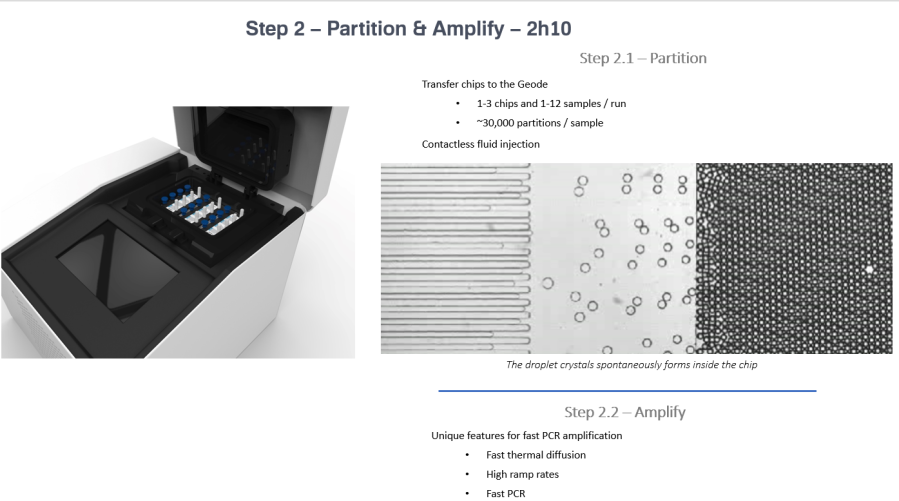
More information about the Naica ™ Crystal Digital PCR workflow can be found at (https://www.stillatechnologies.com/naica-system-workflow/). Tech support at Stilla Technologies are extremely helpful and knowledgeable and can offer additional support for setting up new protocols as well as helping to interpret data and troubleshoot problems (we have had a few – we’re still learning!). If you have a project that you think would benefit from dPCR, then why not contact us at the Genetics Core (https://www.ed.ac.uk/clinical-research-facility/core-services/genetics).
Tammy is a part-time research technician at the ECRF Genetics Core and a full-time Mum of 2 boys, currently specialising in steam trains and Lego Star Wars. As a member of the ECRF Sustainability committee she is committed to ‘greening’ the workplace and improving staff well-being.
